Curate datasets¶
Data curation with LaminDB ensures your datasets are validated, standardized, and queryable. This guide shows you how to transform messy, real-world data into clean, annotated datasets.
Curating a dataset with LaminDB means three things:
✅ Validate that the dataset matches a desired schema
🔧 Standardize the dataset (e.g., by fixing typos, mapping synonyms) or update registries if validation fails
🏷️ Annotate the dataset by linking it against metadata entities so that it becomes queryable
In this guide we’ll curate common data structures. Here is a guide for the underlying low-level API.
Note: If you know either pydantic or pandera, here is an FAQ that compares LaminDB with both of these tools.
# pip install 'lamindb[bionty]'
!lamin init --storage ./test-curate --modules bionty
Show code cell output
→ initialized lamindb: testuser1/test-curate
import lamindb as ln
ln.track("MCeA3reqZG2e")
Show code cell output
→ connected lamindb: testuser1/test-curate
→ created Transform('MCeA3reqZG2e0000'), started new Run('GewHo5l7...') at 2025-07-17 16:12:24 UTC
→ notebook imports: lamindb==1.8.0
Schema design patterns¶
A Schema in LaminDB is a specification that defines the expected structure, data types, and validation rules for a dataset.
Schemas ensure data consistency by defining:
What features (columns/dimensions) should exist in your data
What data types those features should have
What values are valid for categorical features
Which features are required vs optional
Key components of a schema:
schema = ln.Schema(
name="experiment_schema", # Human-readable name
features=[ # Required features
ln.Feature(name="cell_type", dtype=bt.CellType),
ln.Feature(name="treatment", dtype=str),
],
flexible=True, # Allow additional features?
otype="DataFrame" # Object type (DataFrame, AnnData, etc.)
)
For Complex Data Structures:
# AnnData with multiple "slots"
adata_schema = ln.Schema(
otype="AnnData",
slots={
"obs": cell_metadata_schema, # Cell annotations
"var.T": gene_id_schema # Gene features
}
)
Before diving into curation, let’s understand the different schema approaches and when to use each one. Think of schemas as rules that define what valid data should look like.
Flexible schema¶
Validates against any features in your existing registries.
import lamindb as ln
schema = ln.Schema(name="valid_features", itype=ln.Feature).save()
Minimal required schema¶
If we’d like to curate the dataframe with a minimal set of required columns, we can use the following schema.
import lamindb as ln
schema = ln.Schema(
name="Mini immuno schema",
features=[
ln.Feature.get(name="perturbation"),
ln.Feature.get(name="cell_type_by_model"),
ln.Feature.get(name="assay_oid"),
ln.Feature.get(name="donor"),
ln.Feature.get(name="concentration"),
ln.Feature.get(name="treatment_time_h"),
],
flexible=True, # _additional_ columns in a dataframe are validated & annotated
).save()
DataFrame¶
Step 1: Load and examine your data¶
We’ll be working with the mini immuno dataset:
df = ln.core.datasets.mini_immuno.get_dataset1(
with_cell_type_synonym=True, with_cell_type_typo=True
)
df
Show code cell output
| ENSG00000153563 | ENSG00000010610 | ENSG00000170458 | perturbation | sample_note | cell_type_by_expert | cell_type_by_model | assay_oid | concentration | treatment_time_h | donor | donor_ethnicity | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sample1 | 1 | 3 | 5 | DMSO | was ok | B-cell | B cell | EFO:0008913 | 0.1% | 24 | D0001 | [Chinese, Singaporean Chinese] |
| sample2 | 2 | 4 | 6 | IFNG | looks naah | CD8-pos alpha-beta T cell | T cell | EFO:0008913 | 200 nM | 24 | D0002 | [Chinese, Han Chinese] |
| sample3 | 3 | 5 | 7 | DMSO | pretty! 🤩 | CD8-pos alpha-beta T cell | T cell | EFO:0008913 | 0.1% | 6 | None | [Chinese] |
Step 2: Set up your metadata registries¶
Before creating a schema, ensure your registries have the right features and labels:
import lamindb as ln
import bionty as bt
# define valid labels
perturbation_type = ln.ULabel(name="Perturbation", is_type=True).save()
ln.ULabel(name="DMSO", type=perturbation_type).save()
ln.ULabel(name="IFNG", type=perturbation_type).save()
bt.CellType.from_source(name="B cell").save()
bt.CellType.from_source(name="T cell").save()
# define valid features
ln.Feature(name="perturbation", dtype=perturbation_type).save()
ln.Feature(name="cell_type_by_expert", dtype=bt.CellType).save()
ln.Feature(name="cell_type_by_model", dtype=bt.CellType).save()
ln.Feature(name="assay_oid", dtype=bt.ExperimentalFactor.ontology_id).save()
ln.Feature(name="concentration", dtype=str).save()
ln.Feature(name="treatment_time_h", dtype="num", coerce_dtype=True).save()
ln.Feature(name="donor", dtype=str, nullable=True).save()
ln.Feature(name="donor_ethnicity", dtype=list[bt.Ethnicity]).save()
Step 3: Create your schema¶
schema = ln.core.datasets.mini_immuno.define_mini_immuno_schema_flexible()
schema.describe()
Schema ├── .uid = 'q6ycgnZiLeD5JG3B' ├── .name = 'Mini immuno schema' ├── .itype = 'Feature' ├── .ordered_set = False ├── .maximal_set = False ├── .minimal_set = True ├── .created_by = testuser1 (Test User1) ├── .created_at = 2025-07-17 16:12:28 └── Feature • 6 └── name dtype optional nullab… coerce_dtype default_val… perturbation cat[ULabel[Perturbation]] ✗ ✓ ✗ unset cell_type_by_mod… cat[bionty.CellType] ✗ ✓ ✗ unset assay_oid cat[bionty.ExperimentalFactor.ontology_i… ✗ ✓ ✗ unset donor str ✗ ✓ ✗ unset concentration str ✗ ✓ ✗ unset treatment_time_h num ✗ ✓ ✓ unset
Step 4: Initialize Curator and first Validation¶
If you expect the validation to pass, can directly register an artifact by providing the schema:
artifact = ln.Artifact.from_df(df, key="examples/my_curated_dataset.parquet", schema=schema).save()
The validate() method validates that your dataset adheres to the criteria defined by the schema. It identifies which values are already validated (exist in the registries) and which are potentially problematic (do not yet exist in our registries).
try:
curator = ln.curators.DataFrameCurator(df, schema)
curator.validate()
except ln.errors.ValidationError as error:
print(error)
Show code cell output
! 4 terms not validated in feature 'columns': 'ENSG00000153563', 'ENSG00000010610', 'ENSG00000170458', 'sample_note'
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('columns')
! 2 terms not validated in feature 'cell_type_by_expert': 'B-cell', 'CD8-pos alpha-beta T cell'
1 synonym found: "B-cell" → "B cell"
→ curate synonyms via: .standardize("cell_type_by_expert")
for remaining terms:
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('cell_type_by_expert')
2 terms not validated in feature 'cell_type_by_expert': 'B-cell', 'CD8-pos alpha-beta T cell'
1 synonym found: "B-cell" → "B cell"
→ curate synonyms via: .standardize("cell_type_by_expert")
for remaining terms:
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('cell_type_by_expert')
Step 5: Fix Validation Issues¶
# check the non-validated terms
curator.cat.non_validated
Show code cell output
{'cell_type_by_expert': ['B-cell', 'CD8-pos alpha-beta T cell']}
For cell_type_by_expert, we saw 2 terms are not validated.
First, let’s standardize synonym “B-cell” as suggested
curator.cat.standardize("cell_type_by_expert")
# now we have only one non-validated cell type left
curator.cat.non_validated
Show code cell output
{'cell_type_by_expert': ['CD8-pos alpha-beta T cell']}
For “CD8-pos alpha-beta T cell”, let’s understand which cell type in the public ontology might be the actual match.
# to check the correct spelling of categories, pass `public=True` to get a lookup object from public ontologies
# use `lookup = curator.cat.lookup()` to get a lookup object of existing records in your instance
lookup = curator.cat.lookup(public=True)
lookup
Show code cell output
Lookup objects from the public:
.perturbation
.cell_type_by_expert
.cell_type_by_model
.assay_oid
.donor_ethnicity
.columns
Example:
→ categories = curator.lookup()["cell_type"]
→ categories.alveolar_type_1_fibroblast_cell
To look up public ontologies, use .lookup(public=True)
# here is an example for the "cell_type" column
cell_types = lookup["cell_type_by_expert"]
cell_types.cd8_positive_alpha_beta_t_cell
Show code cell output
CellType(ontology_id='CL:0000625', name='CD8-positive, alpha-beta T cell', definition='A T Cell Expressing An Alpha-Beta T Cell Receptor And The Cd8 Coreceptor.', synonyms='CD8-positive, alpha-beta T-cell|CD8-positive, alpha-beta T lymphocyte|CD8-positive, alpha-beta T-lymphocyte', parents=array(['CL:0000791'], dtype=object))
# fix the cell type name
df["cell_type_by_expert"] = df["cell_type_by_expert"].cat.rename_categories(
{"CD8-pos alpha-beta T cell": cell_types.cd8_positive_alpha_beta_t_cell.name}
)
For perturbation, we want to add the new values: “DMSO”, “IFNG”
# this adds perturbations that were _not_ validated
curator.cat.add_new_from("perturbation")
# validate again
curator.validate()
Show code cell output
! 4 terms not validated in feature 'columns': 'ENSG00000153563', 'ENSG00000010610', 'ENSG00000170458', 'sample_note'
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('columns')
Step 6: Save Your Curated Dataset¶
artifact = curator.save_artifact(key="examples/my_curated_dataset.parquet")
artifact.describe()
Show code cell output
Artifact .parquet · DataFrame · dataset ├── General │ ├── key: examples/my_curated_dataset.parquet │ ├── uid: lak2CWzoc9ezJBu80000 hash: kQSstgz6tk5ug4-rq8yz0A │ ├── size: 9.6 KB transform: curate.ipynb │ ├── space: all branch: main │ ├── created_by: testuser1 (Test User1) created_at: 2025-07-17 16:12:33 │ ├── n_observations: 3 │ └── storage path: /home/runner/work/lamindb/lamindb/docs/test-curate/.lamindb/lak2CWzoc9ezJBu80000.parquet ├── Dataset features │ └── columns • 8 [Feature] │ assay_oid cat[bionty.ExperimentalFactor.on… single-cell RNA sequencing │ cell_type_by_expert cat[bionty.CellType] B cell, CD8-positive, alpha-beta T cell │ cell_type_by_model cat[bionty.CellType] B cell, T cell │ donor_ethnicity list[cat[bionty.Ethnicity]] Chinese, Han Chinese, Singaporean Chine… │ perturbation cat[ULabel[Perturbation]] DMSO, IFNG │ concentration str │ treatment_time_h num │ donor str └── Labels └── .cell_types bionty.CellType B cell, T cell, CD8-positive, alpha-bet… .experimental_factors bionty.ExperimentalFactor single-cell RNA sequencing .ethnicities bionty.Ethnicity Chinese, Singaporean Chinese, Han Chine… .ulabels ULabel DMSO, IFNG
AnnData¶
AnnData like all other data structures that follow is a composite structure that stores different arrays in different slots.
Allow a flexible schema¶
We can also allow a flexible schema for an AnnData and only require that it’s indexed with Ensembl gene IDs.
import lamindb as ln
ln.core.datasets.mini_immuno.define_features_labels()
adata = ln.core.datasets.mini_immuno.get_dataset1(otype="AnnData")
schema = ln.examples.schemas.anndata_ensembl_gene_ids_and_valid_features_in_obs()
artifact = ln.Artifact.from_anndata(
adata, key="examples/mini_immuno.h5ad", schema=schema
).save()
artifact.describe()
Let’s run the script.
!python scripts/curate_anndata_flexible.py
Show code cell output
→ connected lamindb: testuser1/test-curate
→ returning existing ULabel record with same name: 'Perturbation'
→ returning existing ULabel record with same name: 'DMSO'
→ returning existing ULabel record with same name: 'IFNG'
→ returning existing Feature record with same name: 'perturbation'
→ returning existing Feature record with same name: 'cell_type_by_expert'
→ returning existing Feature record with same name: 'cell_type_by_model'
→ returning existing Feature record with same name: 'assay_oid'
→ returning existing Feature record with same name: 'concentration'
→ returning existing Feature record with same name: 'treatment_time_h'
→ returning existing Feature record with same name: 'donor'
→ returning existing Feature record with same name: 'donor_ethnicity'
→ connected lamindb: testuser1/test-curate
→ connected lamindb: testuser1/test-curate
! no run & transform got linked, call `ln.track()` & re-run
! 1 term not validated in feature 'columns' in slot 'obs': 'sample_note'
→ fix typos, remove non-existent values, or save terms via: curator.slots['obs'].cat.add_new_from('columns')
Artifact .h5ad · AnnData · dataset
├── General
│ ├── key: examples/mini_immuno.h5ad
│ ├── uid: nDCdF5ER3npirXCV0000 hash: FB3CeMjmg1ivN6HDy6wsSg
│ ├── size: 30.9 KB transform: none
│ ├── space: all branch: main
│ ├── created_by: testuser1 (Test User1) created_at: 2025-07-17 16:12:47
│ ├── n_observations: 3
│ └── storage path:
│ /home/runner/work/lamindb/lamindb/docs/test-curate/.lamindb/nDCdF5ER3npi
│ rXCV0000.h5ad
├── Dataset features
│ ├── obs • 7 [Feature]
│ │ assay_oid cat[bionty.Experiment… single-cell RNA sequencing
│ │ cell_type_by_expe… cat[bionty.CellType] B cell, CD8-positive, alpha…
│ │ cell_type_by_model cat[bionty.CellType] B cell, T cell
│ │ perturbation cat[ULabel[Perturbati… DMSO, IFNG
│ │ concentration str
│ │ treatment_time_h num
│ │ donor str
│ └── var.T • 3 [bionty.Gene.ensembl_…
│ CD8A num
│ CD4 num
│ CD14 num
└── Labels
└── .cell_types bionty.CellType B cell, T cell, CD8-positiv…
.experimental_fac… bionty.ExperimentalFa… single-cell RNA sequencing
.ulabels ULabel DMSO, IFNG
Under-the-hood, this used the following schema:
import lamindb as ln
import bionty as bt
obs_schema = ln.examples.schemas.valid_features()
varT_schema = ln.Schema(
name="valid_ensembl_gene_ids", itype=bt.Gene.ensembl_gene_id
).save()
schema = ln.Schema(
name="anndata_ensembl_gene_ids_and_valid_features_in_obs",
otype="AnnData",
slots={"obs": obs_schema, "var.T": varT_schema},
).save()
This schema tranposes the var DataFrame during curation, so that one validates and annotates the var.T schema, i.e., [ENSG00000153563, ENSG00000010610, ENSG00000170458].
If one doesn’t transpose, one would annotate with the schema of var, i.e., [gene_symbol, gene_type].
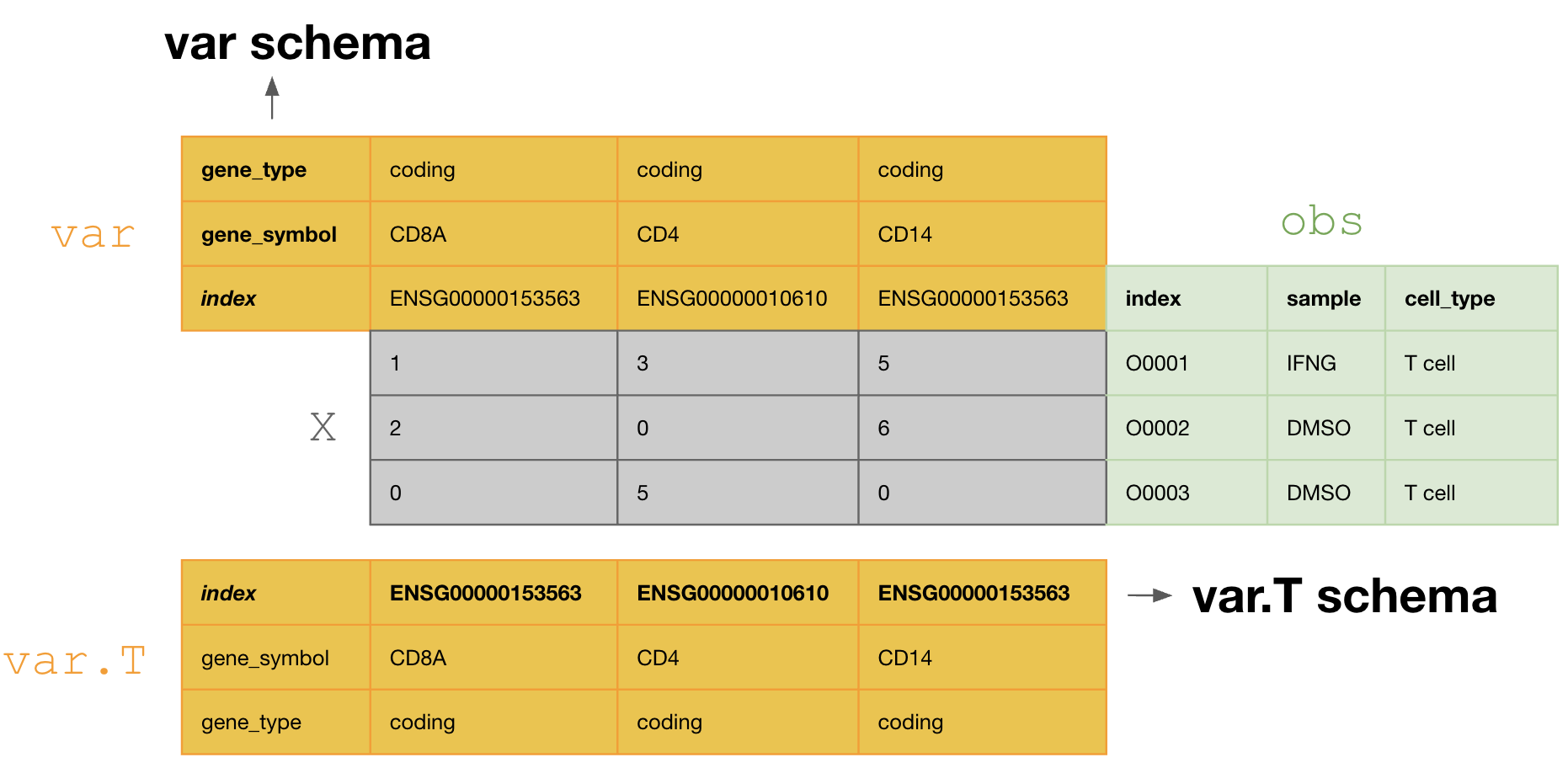
Fix validation issues¶
import lamindb as ln
adata = ln.core.datasets.mini_immuno.get_dataset1(
with_gene_typo=True, with_cell_type_typo=True, otype="AnnData"
)
adata
Show code cell output
AnnData object with n_obs × n_vars = 3 × 3
obs: 'perturbation', 'sample_note', 'cell_type_by_expert', 'cell_type_by_model', 'assay_oid', 'concentration', 'treatment_time_h', 'donor'
uns: 'temperature', 'experiment', 'date_of_study', 'study_note'
Show code cell content
schema = ln.examples.schemas.anndata_ensembl_gene_ids_and_valid_features_in_obs()
schema.describe()
Schema(uid='0000000000000002', name='anndata_ensembl_gene_ids_and_valid_features_in_obs', n=-1, is_type=False, itype='Composite', otype='AnnData', dtype='num', hash='GTxxM36n9tocphLfdbNt9g', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, created_at=2025-07-17 16:12:42 UTC)
obs: Schema(uid='0000000000000000', name='valid_features', n=-1, is_type=False, itype='Feature', hash='kMi7B_N88uu-YnbTLDU-DA', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, created_at=2025-07-17 16:12:42 UTC)
var.T: Schema(uid='0000000000000001', name='valid_ensembl_gene_ids', n=-1, is_type=False, itype='bionty.Gene.ensembl_gene_id', dtype='num', hash='1gocc_TJ1RU2bMwDRK-WUA', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, created_at=2025-07-17 16:12:42 UTC)
Check the slots of a schema:
schema.slots
Show code cell output
{'obs': Schema(uid='0000000000000000', name='valid_features', n=-1, is_type=False, itype='Feature', hash='kMi7B_N88uu-YnbTLDU-DA', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, created_at=2025-07-17 16:12:42 UTC),
'var.T': Schema(uid='0000000000000001', name='valid_ensembl_gene_ids', n=-1, is_type=False, itype='bionty.Gene.ensembl_gene_id', dtype='num', hash='1gocc_TJ1RU2bMwDRK-WUA', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, created_at=2025-07-17 16:12:42 UTC)}
curator = ln.curators.AnnDataCurator(adata, schema)
try:
curator.validate()
except ln.errors.ValidationError as error:
print(error)
Show code cell output
! 1 term not validated in feature 'columns' in slot 'obs': 'sample_note'
→ fix typos, remove non-existent values, or save terms via: curator.slots['obs'].cat.add_new_from('columns')
! 1 term not validated in feature 'cell_type_by_expert' in slot 'obs': 'CD8-pos alpha-beta T cell'
→ fix typos, remove non-existent values, or save terms via: curator.slots['obs'].cat.add_new_from('cell_type_by_expert')
1 term not validated in feature 'cell_type_by_expert' in slot 'obs': 'CD8-pos alpha-beta T cell'
→ fix typos, remove non-existent values, or save terms via: curator.slots['obs'].cat.add_new_from('cell_type_by_expert')
As above, we leverage a lookup object with valid cell types to find the correct name.
valid_cell_types = curator.slots["obs"].cat.lookup()["cell_type_by_expert"]
adata.obs["cell_type_by_expert"] = adata.obs[
"cell_type_by_expert"
].cat.rename_categories(
{"CD8-pos alpha-beta T cell": valid_cell_types.cd8_positive_alpha_beta_t_cell.name}
)
The validated AnnData can be subsequently saved as an Artifact:
adata.obs.columns
Index(['perturbation', 'sample_note', 'cell_type_by_expert',
'cell_type_by_model', 'assay_oid', 'concentration', 'treatment_time_h',
'donor'],
dtype='object')
curator.slots["var.T"].cat.add_new_from("columns")
! using default organism = human
! 1 term not validated in feature 'columns' in slot 'var.T': 'GeneTypo'
→ fix typos, remove non-existent values, or save terms via: curator.slots['var.T'].cat.add_new_from('columns')
curator.validate()
! 1 term not validated in feature 'columns' in slot 'obs': 'sample_note'
→ fix typos, remove non-existent values, or save terms via: curator.slots['obs'].cat.add_new_from('columns')
artifact = curator.save_artifact(key="examples/my_curated_anndata.h5ad")
Show code cell output
→ returning existing schema with same hash: Schema(uid='j9sngdR1IZjAGSNW', n=7, is_type=False, itype='Feature', hash='QTvcEOp8pyRD_oACXWxP3Q', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, created_at=2025-07-17 16:12:47 UTC)
Access the schema for each slot:
artifact.features.slots
Show code cell output
{'obs': Schema(uid='j9sngdR1IZjAGSNW', n=7, is_type=False, itype='Feature', hash='QTvcEOp8pyRD_oACXWxP3Q', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, created_at=2025-07-17 16:12:47 UTC),
'var.T': Schema(uid='KtDTRtmN6sdMVh51', n=3, is_type=False, itype='bionty.Gene.ensembl_gene_id', dtype='num', hash='8e68Zm15DA4DuC39LJr6JA', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, run_id=1, created_at=2025-07-17 16:13:04 UTC)}
The saved artifact has been annotated with validated features and labels:
artifact.describe()
Show code cell output
Artifact .h5ad · AnnData · dataset ├── General │ ├── key: examples/my_curated_anndata.h5ad │ ├── uid: gMPARLhT1EAkD0Kn0000 hash: yeNWx0-dOGGkANQbocU4Sg │ ├── size: 30.9 KB transform: curate.ipynb │ ├── space: all branch: main │ ├── created_by: testuser1 (Test User1) created_at: 2025-07-17 16:13:04 │ ├── n_observations: 3 │ └── storage path: /home/runner/work/lamindb/lamindb/docs/test-curate/.lamindb/gMPARLhT1EAkD0Kn0000.h5ad ├── Dataset features │ ├── obs • 7 [Feature] │ │ assay_oid cat[bionty.ExperimentalFactor.on… single-cell RNA sequencing │ │ cell_type_by_expert cat[bionty.CellType] B cell, CD8-positive, alpha-beta T cell │ │ cell_type_by_model cat[bionty.CellType] B cell, T cell │ │ perturbation cat[ULabel[Perturbation]] DMSO, IFNG │ │ concentration str │ │ treatment_time_h num │ │ donor str │ └── var.T • 3 [bionty.Gene.ensembl_gene_id] │ CD8A num │ CD4 num └── Labels └── .cell_types bionty.CellType B cell, T cell, CD8-positive, alpha-bet… .experimental_factors bionty.ExperimentalFactor single-cell RNA sequencing .ulabels ULabel DMSO, IFNG
MuData¶
import lamindb as ln
import bionty as bt
# define the global obs schema
obs_schema = ln.Schema(
name="mudata_papalexi21_subset_obs_schema",
features=[
ln.Feature(name="perturbation", dtype="cat[ULabel[Perturbation]]").save(),
ln.Feature(name="replicate", dtype="cat[ULabel[Replicate]]").save(),
],
).save()
# define the ['rna'].obs schema
obs_schema_rna = ln.Schema(
name="mudata_papalexi21_subset_rna_obs_schema",
features=[
ln.Feature(name="nCount_RNA", dtype=int).save(),
ln.Feature(name="nFeature_RNA", dtype=int).save(),
ln.Feature(name="percent.mito", dtype=float).save(),
],
).save()
# define the ['hto'].obs schema
obs_schema_hto = ln.Schema(
name="mudata_papalexi21_subset_hto_obs_schema",
features=[
ln.Feature(name="nCount_HTO", dtype=int).save(),
ln.Feature(name="nFeature_HTO", dtype=int).save(),
ln.Feature(name="technique", dtype=bt.ExperimentalFactor).save(),
],
).save()
# define ['rna'].var schema
var_schema_rna = ln.Schema(
name="mudata_papalexi21_subset_rna_var_schema",
itype=bt.Gene.symbol,
dtype=float,
).save()
# define composite schema
mudata_schema = ln.Schema(
name="mudata_papalexi21_subset_mudata_schema",
otype="MuData",
slots={
"obs": obs_schema,
"rna:obs": obs_schema_rna,
"hto:obs": obs_schema_hto,
"rna:var": var_schema_rna,
},
).save()
# curate a MuData
mdata = ln.core.datasets.mudata_papalexi21_subset()
bt.settings.organism = "human" # set the organism to map gene symbols
curator = ln.curators.MuDataCurator(mdata, mudata_schema)
artifact = curator.save_artifact(key="examples/mudata_papalexi21_subset.h5mu")
assert artifact.schema == mudata_schema
!python scripts/curate_mudata.py
Show code cell output
→ connected lamindb: testuser1/test-curate
→ returning existing Feature record with same name: 'perturbation'
! you are trying to create a record with name='nFeature_HTO' but a record with similar name exists: 'nFeature_RNA'. Did you mean to load it?
! auto-transposed `var` for backward compat, please indicate transposition in the schema definition by calling out `.T`: slots={'var.T': itype=bt.Gene.ensembl_gene_id}
! 37 terms not validated in feature 'columns': 'adt:G2M.Score', 'adt:HTO_classification', 'adt:MULTI_ID', 'adt:NT', 'adt:Phase', 'adt:S.Score', 'adt:gene_target', 'adt:guide_ID', 'adt:orig.ident', 'adt:percent.mito', 'adt:perturbation', 'adt:replicate', 'hto:G2M.Score', 'hto:HTO_classification', 'hto:MULTI_ID', 'hto:NT', 'hto:Phase', 'hto:S.Score', 'hto:gene_target', 'hto:guide_ID', ...
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('columns')
! 2 terms not validated in feature 'perturbation': 'Perturbed', 'NT'
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('perturbation')
→ a valid label for subtype 'Perturbation' has to be one of ['DMSO', 'IFNG']
lamindb.models.ulabel.ULabel.DoesNotExist
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/home/runner/work/lamindb/lamindb/docs/scripts/curate_mudata.py", line 57, in <module>
artifact = curator.save_artifact(key="examples/mudata_papalexi21_subset.h5mu")
File "/home/runner/work/lamindb/lamindb/lamindb/curators/core.py", line 335, in save_artifact
self.validate()
~~~~~~~~~~~~~^^
File "/home/runner/work/lamindb/lamindb/lamindb/curators/core.py", line 320, in validate
curator.validate()
~~~~~~~~~~~~~~~~^^
File "/home/runner/work/lamindb/lamindb/lamindb/curators/core.py", line 658, in validate
self._cat_manager_validate()
~~~~~~~~~~~~~~~~~~~~~~~~~~^^
File "/home/runner/work/lamindb/lamindb/lamindb/curators/core.py", line 642, in _cat_manager_validate
self.cat.validate()
~~~~~~~~~~~~~~~~~^^
File "/home/runner/work/lamindb/lamindb/lamindb/curators/core.py", line 1510, in validate
cat_vector.validate()
~~~~~~~~~~~~~~~~~~~^^
File "/home/runner/work/lamindb/lamindb/lamindb/curators/core.py", line 1352, in validate
self._validated, self._non_validated = self._add_validated()
~~~~~~~~~~~~~~~~~~~^^
File "/home/runner/work/lamindb/lamindb/lamindb/curators/core.py", line 1169, in _add_validated
type_record = registry.get(name=self._subtype_str)
File "/home/runner/work/lamindb/lamindb/lamindb/models/sqlrecord.py", line 464, in get
return QuerySet(model=cls).get(idlike, **expressions)
~~~~~~~~~~~~~~~~~~~~~~~^^^^^^^^^^^^^^^^^^^^^^^
File "/home/runner/work/lamindb/lamindb/lamindb/models/query_set.py", line 873, in get
record = get(self, idlike, **expressions)
File "/home/runner/work/lamindb/lamindb/lamindb/models/query_set.py", line 226, in get
raise registry.DoesNotExist from registry.DoesNotExist
lamindb.models.ulabel.ULabel.DoesNotExist
SpatialData¶
import lamindb as ln
import bionty as bt
attrs_schema = ln.Schema(
features=[
ln.Feature(name="bio", dtype=dict).save(),
ln.Feature(name="tech", dtype=dict).save(),
],
).save()
sample_schema = ln.Schema(
features=[
ln.Feature(name="disease", dtype=bt.Disease, coerce_dtype=True).save(),
ln.Feature(
name="developmental_stage",
dtype=bt.DevelopmentalStage,
coerce_dtype=True,
).save(),
],
).save()
tech_schema = ln.Schema(
features=[
ln.Feature(name="assay", dtype=bt.ExperimentalFactor, coerce_dtype=True).save(),
],
).save()
obs_schema = ln.Schema(
features=[
ln.Feature(name="sample_region", dtype="str").save(),
],
).save()
# Schema enforces only registered Ensembl Gene IDs are valid (maximal_set=True)
varT_schema = ln.Schema(itype=bt.Gene.ensembl_gene_id, maximal_set=True).save()
sdata_schema = ln.Schema(
name="spatialdata_blobs_schema",
otype="SpatialData",
slots={
"attrs:bio": sample_schema,
"attrs:tech": tech_schema,
"attrs": attrs_schema,
"tables:table:obs": obs_schema,
"tables:table:var.T": varT_schema,
},
).save()
!python scripts/define_schema_spatialdata.py
Show code cell output
→ connected lamindb: testuser1/test-curate
! you are trying to create a record with name='tech' but a record with similar name exists: 'technique'. Did you mean to load it?
! you are trying to create a record with name='assay' but a record with similar name exists: 'assay_oid'. Did you mean to load it?
import lamindb as ln
spatialdata = ln.core.datasets.spatialdata_blobs()
sdata_schema = ln.Schema.get(name="spatialdata_blobs_schema")
curator = ln.curators.SpatialDataCurator(spatialdata, sdata_schema)
try:
curator.validate()
except ln.errors.ValidationError:
pass
spatialdata.tables["table"].var.drop(index="ENSG00000999999", inplace=True)
# validate again (must pass now) and save artifact
artifact = ln.Artifact.from_spatialdata(
spatialdata, key="examples/spatialdata1.zarr", schema=sdata_schema
).save()
artifact.describe()
!python scripts/curate_spatialdata.py
Show code cell output
→ connected lamindb: testuser1/test-curate
/opt/hostedtoolcache/Python/3.13.5/x64/lib/python3.13/site-packages/xarray_schema/__init__.py:1: UserWarning: pkg_resources is deprecated as an API. See https://setuptools.pypa.io/en/latest/pkg_resources.html. The pkg_resources package is slated for removal as early as 2025-11-30. Refrain from using this package or pin to Setuptools<81.
from pkg_resources import DistributionNotFound, get_distribution
/opt/hostedtoolcache/Python/3.13.5/x64/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:532: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left = partial(_left_join_spatialelement_table)
/opt/hostedtoolcache/Python/3.13.5/x64/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:533: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
left_exclusive = partial(_left_exclusive_join_spatialelement_table)
/opt/hostedtoolcache/Python/3.13.5/x64/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:534: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
inner = partial(_inner_join_spatialelement_table)
/opt/hostedtoolcache/Python/3.13.5/x64/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:535: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right = partial(_right_join_spatialelement_table)
/opt/hostedtoolcache/Python/3.13.5/x64/lib/python3.13/site-packages/spatialdata/_core/query/relational_query.py:536: FutureWarning: functools.partial will be a method descriptor in future Python versions; wrap it in enum.member() if you want to preserve the old behavior
right_exclusive = partial(_right_exclusive_join_spatialelement_table)
/opt/hostedtoolcache/Python/3.13.5/x64/lib/python3.13/site-packages/spatialdata/models/models.py:1144: UserWarning: Converting `region_key: region` to categorical dtype.
return convert_region_column_to_categorical(adata)
! 1 term not validated in feature 'columns' in slot 'attrs': 'random_int'
→ fix typos, remove non-existent values, or save terms via: curator.slots['attrs'].cat.add_new_from('columns')
! 2 terms not validated in feature 'columns' in slot 'tables:table:obs': 'instance_id', 'region'
→ fix typos, remove non-existent values, or save terms via: curator.slots['tables:table:obs'].cat.add_new_from('columns')
! 1 term not validated in feature 'columns' in slot 'tables:table:var.T': 'ENSG00000999999'
→ fix typos, remove non-existent values, or save terms via: curator.slots['tables:table:var.T'].cat.add_new_from('columns')
! no run & transform got linked, call `ln.track()` & re-run
INFO The Zarr backing store has been changed from None the new file path:
/home/runner/.cache/lamindb/eoed5myFanotjILK0000.zarr
! 1 term not validated in feature 'columns' in slot 'attrs': 'random_int'
→ fix typos, remove non-existent values, or save terms via: curator.slots['attrs'].cat.add_new_from('columns')
! 2 terms not validated in feature 'columns' in slot 'tables:table:obs': 'instance_id', 'region'
→ fix typos, remove non-existent values, or save terms via: curator.slots['tables:table:obs'].cat.add_new_from('columns')
→ returning existing schema with same hash: Schema(uid='OaQZPO7o5LMBNAQh', n=2, is_type=False, itype='Feature', hash='mKL5iuJBVJ_atA5fG6KsvA', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, created_at=2025-07-17 16:13:12 UTC)
→ returning existing schema with same hash: Schema(uid='7ZxiTxhYtYE3BDdl', n=1, is_type=False, itype='Feature', hash='zz7raO3Sm-7Ehom6tDiIHA', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, created_at=2025-07-17 16:13:12 UTC)
→ returning existing schema with same hash: Schema(uid='VeLgE0X2592oZdke', n=2, is_type=False, itype='Feature', hash='c9NMaXTqjd8zD4pzXzWvgg', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, created_at=2025-07-17 16:13:12 UTC)
→ returning existing schema with same hash: Schema(uid='FpT3XtO7hVZTfqYv', n=1, is_type=False, itype='Feature', hash='pRiD1iF8DzoQd5cPX2DQhg', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, created_at=2025-07-17 16:13:12 UTC)
Artifact .zarr · SpatialData · dataset
├── General
│ ├── key: examples/spatialdata1.zarr
│ ├── uid: eoed5myFanotjILK0000 hash: LeYCNoxHuOJ_JnWc8oXRPA
│ ├── size: 11.6 MB transform: none
│ ├── space: all branch: main
│ ├── created_by: testuser1 (Test User1) created_at: 2025-07-17 16:13:33
│ ├── n_files: 113
│ └── storage path:
│ /home/runner/work/lamindb/lamindb/docs/test-curate/.lamindb/eoed5myFanot
│ jILK.zarr
├── Dataset features
│ ├── attrs:bio • 2 [Feature]
│ │ developmental_sta… cat[bionty.Developmen… adult stage
│ │ disease cat[bionty.Disease] Alzheimer disease
│ ├── attrs:tech • 1 [Feature]
│ │ assay cat[bionty.Experiment… Visium Spatial Gene Express…
│ ├── attrs • 2 [Feature]
│ │ bio dict
│ │ tech dict
│ ├── tables:table:obs … [Feature]
│ │ sample_region str
│ └── tables:table:var.… [bionty.Gene.ensembl_…
│ BRCA2 num
│ BRAF num
└── Labels
└── .diseases bionty.Disease Alzheimer disease
.experimental_fac… bionty.ExperimentalFa… Visium Spatial Gene Express…
.developmental_st… bionty.DevelopmentalS… adult stage
TiledbsomaExperiment¶
import lamindb as ln
import bionty as bt
import tiledbsoma as soma
import tiledbsoma.io
adata = ln.core.datasets.mini_immuno.get_dataset1(otype="AnnData")
tiledbsoma.io.from_anndata("small_dataset.tiledbsoma", adata, measurement_name="RNA")
obs_schema = ln.Schema(
name="soma_obs_schema",
features=[
ln.Feature(name="cell_type_by_expert", dtype=bt.CellType).save(),
ln.Feature(name="cell_type_by_model", dtype=bt.CellType).save(),
],
).save()
var_schema = ln.Schema(
name="soma_var_schema",
features=[
ln.Feature(name="var_id", dtype=bt.Gene.ensembl_gene_id).save(),
],
coerce_dtype=True,
).save()
soma_schema = ln.Schema(
name="soma_experiment_schema",
otype="tiledbsoma",
slots={
"obs": obs_schema,
"ms:RNA.T": var_schema,
},
).save()
with soma.Experiment.open("small_dataset.tiledbsoma") as experiment:
curator = ln.curators.TiledbsomaExperimentCurator(experiment, soma_schema)
curator.validate()
artifact = curator.save_artifact(
key="examples/soma_experiment.tiledbsoma",
description="SOMA experiment with schema validation",
)
assert artifact.schema == soma_schema
artifact.describe()
!python scripts/curate_soma_experiment.py
Show code cell output
→ connected lamindb: testuser1/test-curate
→ returning existing Feature record with same name: 'cell_type_by_expert'
→ returning existing Feature record with same name: 'cell_type_by_model'
! 1 term not validated in feature 'columns': 'sample_note'
→ fix typos, remove non-existent values, or save terms via: curator.cat.add_new_from('columns')
! no run & transform got linked, call `ln.track()` & re-run
→ returning existing schema with same hash: Schema(uid='j9sngdR1IZjAGSNW', n=7, is_type=False, itype='Feature', hash='QTvcEOp8pyRD_oACXWxP3Q', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, created_at=2025-07-17 16:12:47 UTC)
→ returning existing schema with same hash: Schema(uid='urVdF4LzSvmee34A', name='soma_var_schema', n=1, is_type=False, itype='Feature', hash='ziS-ah8kRjSvXXB6LjGrNQ', minimal_set=True, ordered_set=False, maximal_set=False, branch_id=1, space_id=1, created_by_id=1, created_at=2025-07-17 16:13:37 UTC)
Artifact .tiledbsoma · tiledbsoma · dataset
├── General
│ ├── key: examples/soma_experiment.tiledbsoma
│ ├── description: SOMA experiment with schema validation
│ ├── uid: PrBQrlkXetFhdpdM0000 hash: F-V6Rqa0wRd6BSl0TreB5g
│ ├── size: 23.9 KB transform: none
│ ├── space: all branch: main
│ ├── created_by: testuser1 (Test User1) created_at: 2025-07-17 16:13:39
│ ├── n_files: 68 n_observations: 3
│ └── storage path:
│ /home/runner/work/lamindb/lamindb/docs/test-curate/.lamindb/PrBQrlkXetFh
│ dpdM.tiledbsoma
├── Dataset features
│ ├── obs • 7 [Feature]
│ │ cell_type_by_expe… cat[bionty.CellType] B cell, CD8-positive, alpha…
│ │ cell_type_by_model cat[bionty.CellType] B cell, T cell
│ │ perturbation cat[ULabel[Perturbati…
│ │ assay_oid cat[bionty.Experiment…
│ │ concentration str
│ │ treatment_time_h num
│ │ donor str
│ └── ms:RNA.T • 1 [Feature]
│ var_id cat[bionty.Gene.ensem… CD14, CD4, CD8A
└── Labels
└── .genes bionty.Gene CD8A, CD4, CD14
.cell_types bionty.CellType B cell, T cell, CD8-positiv…
Other data structures¶
If you have other data structures, read: How do I validate & annotate arbitrary data structures?.
Show code cell content
!rm -rf ./test-curate
!rm -rf ./small_dataset.tiledbsoma
!lamin delete --force test-curate
• deleting instance testuser1/test-curate